CLIMB-SUPPORTED TRAINING
CLIMB strives to provide training to microbiologists through regular practical workshops, seminars, and training images on virtual machines. The infrastructure can also be helpful to senior bioinformaticians, or PIs who aim to organize workshops or hands-on training activities to strengthen the UK microbial bioinformatics community.
Last week, on Oct. 18 and 19, Sophie Nixon ran a two-day workshop with a focus on metagenomics on CLIMB servers. Having been in touch with her, we’re proud to host her account of the training activity. Congratulations to Sophie for having successfully run such a workshop, and big thanks for having sent us the feedback that follows!
THE WORKSHOP – FROM SOPHIE NIXON
Metagenomics is a powerful tool that can shed light on the diversity and potential function of microbial communities. It can be particularly valuable in understanding microbial interactions between and survival strategies of microorganisms in extreme environments, where access is often challenging and much of the community uncultivable. The recent decrease in DNA sequencing costs has made this tool more accessible than ever. However, the skills required to analyse the vast amounts of metagenomic sequencing data are lacking.
I was recently invited to deliver a metagenomics data analysis workshop for the Astrobiology group at the Open University. This group studies the microbiology of some of the most extreme environments on Earth, and would like to use metagenomics to better understand the diversity and potential function of these communities. The workshop was aimed at equipping members of the group with the command-line bioinformatics skills required to interrogate metagenomic data. I deliberately designed this workshop to use freely available software packages and cloud-based computational resources to show that you don’t need to be working in a microbiome group with access to an in-house bioinformatician and server to obtain meaningful results from metagenomes. CLIMB was an obvious choice for the workshop, and proved very successful. Since creating my account on CLIMB I have been impressed by the flexibility and easy-to-access computational power offered by virtual machines (VMs), and decided to design the hands-on learn-by-doing workshop using customised CLIMB VMs.
In day 1 of the workshop I delivered a few talks outlining the principles of metagenomics, including the typical workflow of generating and analysing metagenomic data. The participants already had molecular biology experience, so I chose to focus more on the data analysis aspects rather than the DNA extraction and library preparation process. After the talks we spent a few hours making sure everyone could access the VMs we would be using the following day for the data analysis practical. It took a while to make sure everyone was able to log onto their VM via the command line, and transfer files using FileZilla, but we got there in the end and served as an important demonstration of the practical issues that commonly arise in genomic data analysis!
For the hands-on practical on day 2 I set up several identical Custom Ubuntu instances on CLIMB, one for each of the five participants. Onto each VM I transferred the same raw sequencing data files, originating from a water sample from a deep granitic aquifer in the Arctic Circle. The goal of the analysis was to recover good quality draft genomes from the low-diversity microbial community we know exists in this sample from previous amplicon sequencing data. Having recovered genomes, we would begin mining them for indications of who they might represent and what metabolic pathways they are capable of. In addition to the raw data, I also installed Miniconda, a package manager, onto each VM. Participants were to install the tools required for analysis themselves.
In the practical, we followed a workflow that I had put together and tested in the preceding weeks. There are many more tools available than I chose to include in this workflow, including pipelines that automate several steps of this workflow via an intuitive interface. However, the goal was to teach the skills necessary to pick the most suitable tools available to answer the underlying question, and key to this is familiarity with the command line and widely-used reputable tools available.
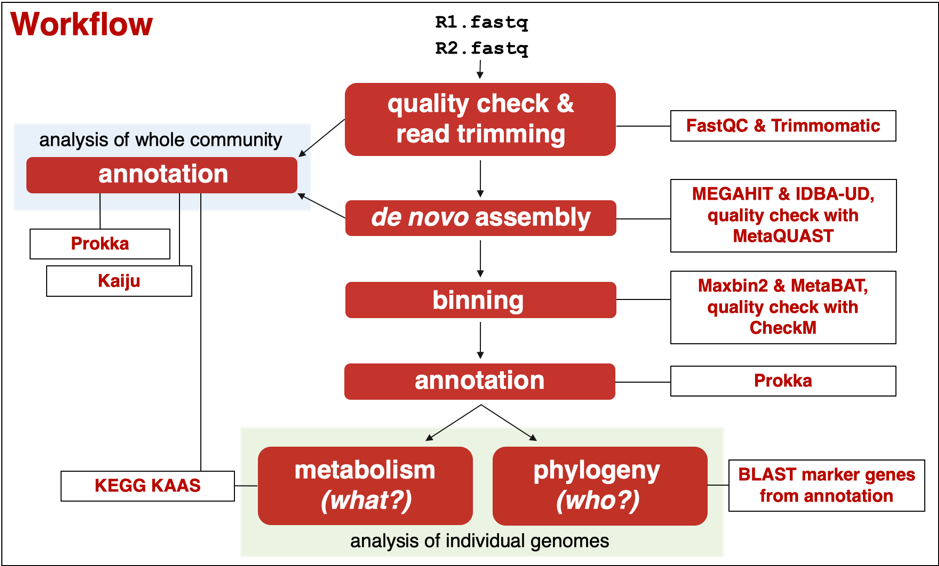
The agenda for the practical was ambitious but by the end of the second day we had recovered good quality draft genomes and had some idea of their metabolic potential and phylogeny. All participants gained valuable command-line experience, and learnt how to set up a customisable VM on CLIMB, connect via a personal computer, install and run relevant software packages on the VM, and transfer files between local and remote drives. They now have the necessary skills required to start analysing their own metagenomic datasets and wow us all about their discoveries of microbial life in Earth’s most extreme environments!
—
Dr Sophie Nixon is a NERC Research Fellow at the School of Earth and Environmental Sciences, University of Manchester. Her research looks to understand the diversity, function and adaptation of microbial life in deep terrestrial habitats, spanning pristine and engineered subsurface environments on Earth, and the potential for life on other planetary bodies. Her current research focuses on microbial life in hydraulically fractured shale environments, and she combines high-pressure subsurface simulation with genomic tools to understand the role of microbiology in shale gas extraction.


